CAGE™ Preparation Kit
CAGE Preparation Kit is a ready-to-use product that includes reagents* and the protocol necessary for the preparation of a CAGE library, which allows the easy introduction of Cap Analysis of Gene Expression, CAGE, into your R&D.
* Note: Reverse Transcriptase is not inclued.
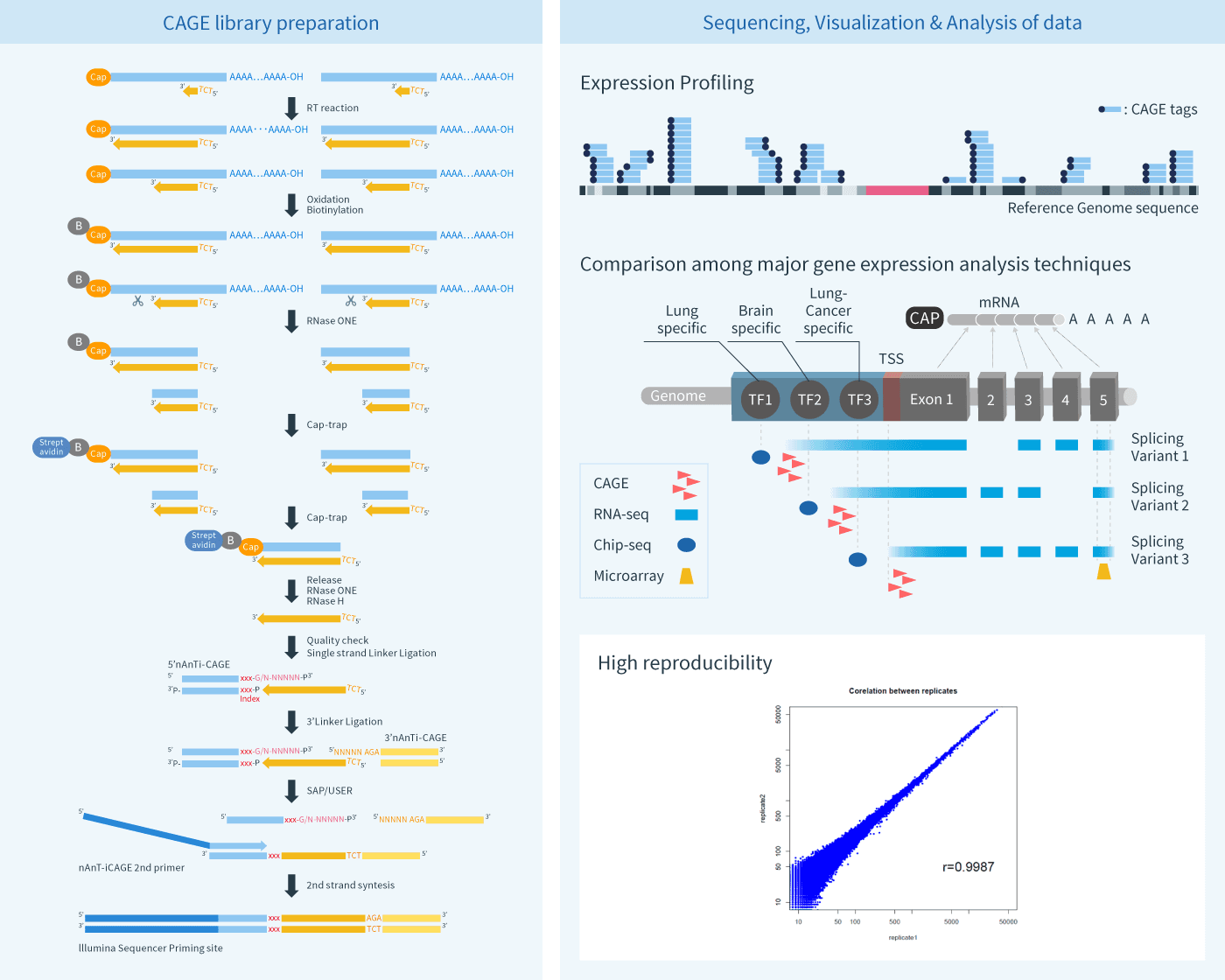
CAGE enables sequencing and mapping of short tags derived from the 5'-end of mRNA, thereby quantifying the frequency of tag sequences. A distinctive feature of CAGE is its capability to accurately identify promoter sites for each transcript and obtain gene expression profiles based on identified promoters.
A sequencing run using one channel on the Illumina Sequencer can yield over 4,000,000 reads per sample. Expression profiles obtained from DeepCAGE (i.e., combination of next-generation sequencing with next-generation expressing profiling) will be powerful tools for various gene expression analyses and genome annotation projects.
What can CAGE offer you?
Unique data
CAGE enables experimental identification of transcriptional start sites and quantification of promoter activity.
Wide range of applications
CAGE enables analysis of transcriptomes from any organism.
Wide dynamic range
CAGE also enables detection of rare transcripts.
Learn more about:
Applications
- Genome-wide gene-expression analysis
- Prediction of promoter sites
- Quantitative mRNA expression profiling
Examples of actual usage of CAGE
- "Functional Annotation of Mouse (FANTOM) project" by RIKEN
- "Encyclopedia of DNA Elements (ENCODE) project" by the National Institute of Health (NIH)
- "Gene Networks project" by the Ministry of Education, Culture, Sports, Science and Technology